Previous Project Information Page
Development of an Improved Chemical Speciation Database
for Processing Emissions of Volatile Organic Compounds for Air Quality
Models
By
William P. L. Carter
College of Engineering, Center for Environmental Research and Technology
(CE-CERT)
University of California, Riverside, CA 92521
Database last updated April 1, 2013
Programs or input files last updated April 30, 2009
Database and documentation superceded on February, 2014. Click here for the current version.
NOTE:
This has been superceded by a completely revised version of the
database and its associated programs, which are available at /~carter/emitdb. This page is provided as a link to the earlier programs and files
Contents
Project Description
Volatile organic compounds (VOCs) differ significantly in their effects
on ozone formation, and these differences need to be represented appropriately
in the airshed models used to predict the effects of changes of emissions
on formation of ozone. This requires appropriate methods to specify the
chemical compositions of the many types of VOCs that are emitted, and appropriate
methods to represent the chemical differences of these compounds in the
models. However, current models and emissions databases have significant
problems in this regard:
-
Most models use highly condensed mechanisms that lump the many different
types of VOCs into a limited number of model species.
-
It is difficult and expensive to re-process emissions if chemical mechanisms
are changed or more model species are added, and do not permit use of the
full capabilities of chemically detailed mechanisms such as SAPRC-99 (Carter,
2000a)
-
The chemical speciation information used in current databases have many
poorly defined and redundant chemical categorizations, and different databases
use inconsistent categorizations. This makes it difficult to implement
new chemical mechanisms or to process emissions for different mechanisms
in a consistent manner.
To address these problems, the University of Houston (UH) funded us to
begin the development of an improved chemical speciation database with
assignments to the current SAPRC-99 mechanism, and the American Chemistry
Council (ACC) funded us to further develop this database and develop the
software and files needed to utilize this database to implement the SAPRC-99
and other chemical mechanisms into the SMOKE emissions processing system.
The UH project has been completed, and the speciation database developed
for this project is described below. The ACC project
has also recently been completed and is also described below.
The final report for this project, titled "Integration of the SAPRC
Chemical Mechanism in the SMOKE Emissions Processor for the CMAQ/Models-3
Airshed Model" is now available (PDF format).
More recently, we have been funded by Pecan
and Associates to make model species classifications for species categories
in the Speciate 4.0 database
that is now under development. This has necessitated the addition of a
number of additional categories, which are now included in the speciation
database developed for the previous projects. As part of this effort, model
speciation assignments were made for the new Carbon Bond '05 mechanism,
so four mechanisms (SAPRC-99, RADM2, CB4, and CB05) are now supported by
the current database.
The emissions speciation database assignments and profile data compiled
for UH, ACC and Speciate 4.0 projects are given in Excel files that can
be downloaded from the links below. However, ultimately
we need the chemical assignments for the speciation profiles to be in a
universally available, web accessible format that can be updated as needed
using state-of-the-art database software.
Development of the Speciation
Database for the University of Houston Project
The development of the speciation database carried out under contract
with the University of Houston has involved the following components:
-
A list of all the chemical categories in the emissions speciation databases
used by the EPA for processing emissions for Models-3 (Gipson,
2001), the EPA’s Speciate 3.2 database, (Speciate,
2001) the Texas Natural Resource Conservation Commission (TNRCC) (Yarwood,
2002), and the California Air Resources Board (CARB) (Allen,
2001) was compiled. The Speciate and California profile databases have
since been updated as part of the ongoing ACC project.
-
Duplicate categories were identified using various methods, a master list
containing unique categories was compiled, and these unique speciation
categories were assigned to the categories used in the various emissions
databases. The resulting database contained 1855 speciation categories
to represent the total of 3639 emissions categories from the 4 databases.
Note that the database included some new categories that were added to
define the compositions of the mixtures, as discussed below.
-
CAS identification numbers were determined for all the speciation categories
where these are well defined and uniquely identify a specific chemical
or mixture, and were used to identify these categories using a "C"+CAS
number designation (e.g., "C74-84-0" for ethane). The Chemfinder.com
and ChemExper.com web sites were
used as the primary means for this purpose, with the SciFinder
Scholar program, which directly accesses the CAS database, being used
for compounds not found on the other databases until that resource became
unavailable as a free resource for academics in early 2002. Chemicals that
do not have CAS numbers that we could determine or mixtures for which CAS
numbers are not assigned or poorly defined were given separate identification
codes based on SAROAD numbers used in EPA or other database (e.g., "S1-43155"
for "isomers of heptadecane", based on the EPA SAROAD number, or "S2-97000"
for "C4-naphthalenes", based on the CARB chemical codes.)
-
Each of the new speciation categories were classified as either individual
compounds, mixtures of isomers ("simple mixtures"), mixtures of compounds
that are not necessarily isomers ("complex mixtures"), which were treated
separately in the new database as described below. The current database
has 1304 individual compounds, 319 simple mixtures, 200 complex mixtures,
and 32 poorly defined classes.
-
For each speciation category identified as an individual compound, the
chemical formula (numbers of each type of atom) was determined and used
as the basis for computing the molecular weight, and attempts were made
to assign these compounds to model species in the SAPRC-99 detailed and
condensed mechanisms and the Carbon Bond 4 (CB4) mechanism. 74% of the
compounds could be assigned to SAPRC-99 detailed model species, 95% to
SAPRC-99 lumped species, and 96% to CB4 groups. The more complete set of
SAPRC-99 lumped species assignments resulted from the fact that a number
of compounds did not correspond to existing SAPRC-99 detailed species but
could be identified as being appropriately lumped with existing SAPRC-99
lumped groups. The CB4 assignments were based primarily on those in the
TNRCC database, with most other CB4 assignments from databases received
previously, though a few new CB4 assignments had to be made for this project.
-
No direct assignments of model species were made to categories representing
mixtures. Instead as many of these as possible were assigned to mixtures
of compounds, whose model species assignments can then be used to determine
the appropriate model species for the mixtures. This approach thus separates
out chemical mechanism assignments from emissions composition assignments,
so each can be updated separately and consistently as improved information
or new mechanisms become available.
-
For each speciation category identified as a mixture of isomers, a distribution
of up to 8 individual compounds was chosen to represent the mixture for
modeling purposes. The mixture was chosen based on either existing assignments
in the SAPRC-99 mechanism or using a representative set of compounds of
this type in the database. Such assignments were made for each of these
"simple mixture" categories.
-
Eventually, most complex mixture categories should be removed from speciation
profiles used in emissions inventories, with the inventory developers providing
their best estimate of the compositions in terms of actual compounds or
mixtures of isomers. However, such categories are important in current
inventories, and at present they cannot be neglected. Therefore, composition
assignments were made for as many of the complex mixture speciation categories
as possible. In most cases, a separate set of "profiles" were used to represent
the compositions of the mixtures, though in a few cases representation
by a single compound or simple mixture was considered to be appropriate.
The representative profiles were taken from existing inventories or estimated
in this work, and their derivations are documented in the database.
-
Some of the 32 poorly defined emissions categories were identified as polymers
or other substances that had no volatility, so these were identified as
such so that their mass would not be counted when making assignments for
emissions. The rest were treated as unspeciated mixtures for emissions
processing purposes.
-
Quality assurance was a major component of this project, and numerous procedures
were used to test for consistency between the various databases and assignments
employed. In some cases corrections had to be made based on interactions
with Environ and others working on existing emissions databases. Nevertheless,
because of the size of the database it is almost certain that at least
some errors or inconsistencies may be present, so further checks and reviews
are advisable.
-
Speciation profiles used by the EPA and the CARB were obtained at the same
time as the speciation categorizations (Gipson, 2001;
Allen,
2001), and the extent to which the current compound and mixture assignments
are sufficient to characterize these profiles was assessed. It was found
that at least 99% of the mass was assigned to SAPRC-99 lumped species in
almost 94% of the EPA profiles, and only one profile, "Solvent Utilization:
Naphthenic Acids," had less than 50% of the mass assigned to detailed or
lumped SAPRC-99 species. A somewhat larger number (~2%) of CARB profiles
were less than 50% assigned, but ~94% of the profiles had over 90% of the
mass assigned to SAPRC-99 model species, with the average for all profiles
being 96%. Information was not available concerning Texas speciation profiles,
so the extent of assignments in that database could not be determined.
However, the coverage is expected to be similar to the EPA data base.
-
Procedures were developed to utilize these assignments to derive the model
species associated with each of the emissions categories used in the various
databases. This includes procedures to produce ASCII files containing the
mechanism assignments for the EPA, CARB, TNRCC, or Speciate 3.1 categories
that can be used by emissions processing programs. However, formatting
these files for use in existing emissions processing programs, or modifying
existing emissions processing programs to utilize these files, was beyond
the scope of the present project.
Summary and Progress to Date on
the American Chemistry Council Project
Project Description and Objectives
Additional work on this speciation database has been initiated as part
of a project being carried out by the University of North Carolina (UNC)
and ourselves at CE-CERT to improve the integration of the SAPRC and other
chemical mechanism into SMOKE modeling system within the Models-3 framework.
This work is being funded by the ACC for the RRWG as described n the Project
Description section, above. The specific objectives
of this project are as follows:
-
Create the speciation database organization and assignments that appropriately
represent relationships between mixtures, actual chemical compounds, and
the model species in SAPRC 99 and other mechanisms. Much of this work has
already been completed for the University of Houston project described
above, but additional work is needed as described above. This task is being
carried out primarily by us at CE-CERT.
-
Develop the software and essential data to update both emissions processing
methods and SMOKE for SAPRC and other mechanisms using the reorganized
speciation database (developed in Task 1) and existing emissions datasets.
This task is being carried out both by CE-CERT and UNC.
-
Test SMOKE with the redesigned speciation database and SAPRC and other
mechanism lumped species as modified by reactivity packets using data available
from one previous modeling application. This is being carried out primarily
by UNC
-
Demonstrate application of reactivity controls to emissions using SAPRC
and the Community Multi-scale Air Quality (CMAQ) modeling system. This
is being carried out primarily by UNC
-
Develop interim report, final report, and peer reviewed journal manuscript.
The following are optional tasks that may be carried out if funding is
available. They would be carried out primarily by UNC.
-
Update SMOKE Tool to support using the SAPRC chemical mechanism with CMAQ.
-
Design and implement a Java interface to prepare SMOKE control packets
(including reactivity packets).
-
Integrate Java interface with Models-3 and SMOKE.
The latter tasks could not be carried out for this project because of limited
resources and time. Given below is a brief summary of the progress to date
on the tasks in this project being carried out at CE-CERT.
Profile Database
We obtained the profile data from Speciate 3.2 (Speciate
2002), and compiled it into an Excel database with updated profiles
we obtained from the California ARB (CARB, 2003),
and profiles we already had from the the EPA (Gipson,
2001), and for Texas (Yarwood, 2002). These
are merged into an Excel file named profdb.xls,
which also includes macros to output the profile data as ASCII files that
can be read by the Profile Preprocessing program discussed below. We have
also obtained updated Texas profiles from Gabriel Cantu of TCEQ, but have
not yet incorporated them. Note that many of these profiles are the same,
though we have not yet determined which are duplicates. This file is available
for downloading and is described in more detail below.
Speciation Processing Programs
The work plan for this project calls for developing a series of FORTRAN
programs so that the speciation database described above can be used when
processing emissions with the SMOKE system. As presently formulated, these
consist of four programs: the Profile Preprocessor (ProfPro), the Speciation
Preprocessor (SpecPro), the Emissions Summary Processor (EmitSum), and
the Mechanism Processor (MechPro). The relationships between these programs
and relevant programs and files in the SMOKE and the CMAQ systems are shown
in Figure 1, below. At the present time, the initial versions of all four
programs have been written and preliminary documentation
has been prepared, and preliminary versions of the programs and their
input and output files are available (see below).
Work on implementing them into the SMOKE and CMAQ systems is underway.
The files and programs discussed in the preliminary documentation are subject
to change as this work proceeds.
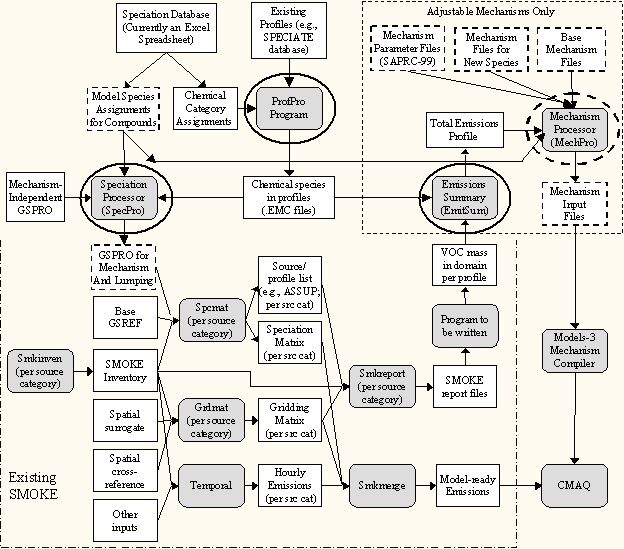
Figure 1. Diagram showing relationships between SMOKE
and the speciation database programs and files. Mechanism-dependent files
are indicated by dashed borders. The programs completed thus far are indicated
with bold ovals, and the program still to be completed for this project
is shown with the dashed oval.
Description of the Speciation
Assignment File
The current set of speciation assignments (which incorporates work on
the ACC project that will be described later) is incorporated in an Excel
2000 file emitdb.xls, which can be downloaded as indicated below. The file
includes a "documentation" sheet that describes in detail the format of
all the other sheets, and also a number of Excel macros that can be used
to process the data or output ASCII files with the mechanism assignments.
Given below is a summary of all the sheets incorporated in the current
database file.
| Documentation |
Summarizes the purpose of each
sheet and describes the columns or data fields they contain. Also gives
the color conventions used throughout the file and reference citations
for additional information on the mechanisms. |
| Parameters and Commands |
Gives the parameters or definitions
used in the various sheets or macros, and also lists the macros used and
gives controls that can be used to run them. See comments in the sheet
for details. |
| Master List |
List of all the emissions classifications
in my current database. Each class should be a unique compound or mixture. |
| Compounds |
Lists the emissions categories
that refer to single compounds (or mixtures of isomers that are not distinguished,
optical isomers), and gives their mechanistic assignments |
| Simple Mixes |
Lists the emissions categories
that refer to mixtures of isomers, and summarizes their assignments to
compounds. (Note that the assignments used are in a separate "Simple Mixes
Asst's" sheet, and the assignments on this sheet are produced by the "CompileSimpMixAsnts"
macro.) |
| Simple Mixes Asst's |
Used to give the assignments of
compounds to the simple mixtures listed in the "Simple Mixes" sheet. One
row is used for each assignment, so multiple rows will be used for mixtures
assigned to more than one compound. |
| Complex Mixtures |
Gives summaries, compositional
assignments, and other information about complex mixtures. Details of assignments
and additional information about complex mixtures are given in separate
sheets as indicated below. |
| Mix Profile Summary |
Gives summary information about
the profiles used to specify the compositions of some of the complex mixtures.
The profiles are associated with the complex mixtures in the "Assignment
of Mwt" column in the "Complex Mixtures" sheet for the mixtures with assignment
type given as "CMP". The composition of the profiles is given in the "Mix
Profile Assignments" sheet, and additional documentation notes are given
in the "Profile Documentation Notes" sheet. |
| Mix Profile Assignments |
Gives composition assigned to
the profiles used to specify the compositions of some of the complex mixtures.
One row is given for each component assigned to a profile, so in general
a profile will have more than one column. |
| Profile Documentation Notes |
Footnotes corresponding to numbers
given in the "notes" column of the mixture profile summary sheet, giving
additional information about the derivation of the composition of the mixture
profiles used. |
| Other Categories |
Lists and gives summary and other
information about the emissions classifications that are poorly defined
or inappropriate or have not yet been categorized. These include (1) compounds
or mixtures that are not appropriate for VOC profiles; (2) Polymers, salts,
or extremely high molecular weight materials; (3) categories that are not
compounds or mixtures, such as elements or CB4 model species; (4) categories
that are designated incorrectly or are too ambiguous to assign; (5) categories
that may be compounds or mixtures of compounds but the compounds' structures
could not be determined or have not yet been determined. (6) categories
that have not yet been classified because they are not present or are extremely
minor constituents in current profiles. |
| Emissions Groups |
Gives the table of SAPRC-99 and
emissions groups and the SAPRC-99 and RADM-2 lumped model species associated
with each. |
| DMS |
Lists SAPRC-99 detailed model
species and gives relevant summary information, including lumped emissions
groups assigned to each. |
| Master Assignments |
Gives the complete mechanism assignments
for all of the emissions categories in the database. All data on the sheet
except the headers are derived using the "MergeAllCmpds" macro. Multiple
rows are given for categories represented by more than one compound, one
per compound used. |
| SAROAD Assignments |
Assigns master emissions database
categories to the SAROAD or chemical codes used in the EPA, California
ARB, TNRCC, and Speciate 3.2 and 4.0 emissions databases. Also gives the
complete mechanism assignments for these categories. The latter are derived
by running the "ProcessSARmodelSpec" macro. |
| Emit Worksheet |
Can be used as an example to derive
detailed and lumped model species assignments for an emissions profile
given in terms of a given type of SAROAD or pseudo-SAROAD categories. |
The file incorporates several macros to output the data in the spreadsheet
into various ASCII files that can be read by the speciation database processing
programs that are being developed for the ACC project discussed
above.
The current version of the speciation assignment file can
be downloaded from ../emitdb/emitdb.xls
Profile Database File
The profile database file contains the profiles in the
Speciate 3.2 database (Speciate, 2002), the
profiles used by the EPA for processing emissions for Models-3 (Gipson,
2001), the Texas Natural Resource Conservation Commission (TNRCC) (Yarwood,
2002), and the profiles provided by the California Air Resources Board
(CARB, 2003). The file contains a "documentation"
sheet that describes the data in the file. Given below is a summary of
the sheets incorporated in the current file.
| Documentation |
Summarizes the purpose of each
sheet and describes the columns or data fields they contain. |
| Parameters |
Gives the parameters and controls
for the macros in this file and also lists and documents the profile types
incorporated in the database. Gives the locations of the profile data (.EMI)
files to be output by the macro in this file and the control to run the
macro. |
| Profile Descriptions |
Lists all the profiles and gives
descriptive and documentation information available for them. Documentation
consists of reference and note numbers that are associated with documentation
text in the "Profile Reference" and "Profile Notes" sheets. |
| Profile Compositions |
Gives the mass fractions of the
chemical categories in the profiles, and also the descriptions of the categories
used in the profile databases as obtained. Note that the categorizations
used are those of the database from which the profiles were obtained, and
differ for each database. The assignments of compounds to these categories
is made in the speciation database incorporated in emitdb.xls. |
| Profile References |
Gives the text associated with
the reference numbers used in the "Profile Descriptions" sheet. |
| Profile Notes |
Gives the text associated with
the note numbers used in the "Profile Descriptions" sheet. |
This Excel file also incorporates a macro that outputs the profile composition
data and available documentation information into the ASCII profile data
(.EMI) files that are used as input to the speciation processing programs
discussed above
The current version of the profile database file,can be
downloaded from ../emitdb/profdb.xls
Additional Work Needed
Although portions of this new database have been provided
to others, the complete database has not been externally reviewed by other
groups or checked for general utility or other problems. However, the available
funding for the project for the University of Houston has been spent, so
the remaining work for this project must be restricted to correcting any
problems or inconsistencies found in the database and completing the final
documentation and report. Additional work to incorporate this into the
SMOKE emissions processing system is underway as part an ACC project
for the RRWG as discussed above.
Even after the current ACC project is completed, this
speciation database must be considered to be a work in progress. Ongoing
work that is needed is summarized below:
-
The assignments of compounds to mixtures need to be reviewed
by experts in emissions speciation. Arbitrary and undocumented assignments
need to be improved, and assignments need to be made for currently unassigned
classes. Note that this is best done by an emissions speciation expert
rather than a chemical mechanism developer.
-
CAS numbers need to be determined or assigned for compounds
for which they could not be determined in current databases. This will
probably require a public agency contracting with CAS to do this.
-
The model species assignments to the individual compounds
need to be independently reviewed and assignments need to be made to more
of the chemical classes that are currently unassigned. This may require
some chemical mechanism development to represent compounds that are not
currently well represented in any mechanism.
-
New compounds will need to be added as needed when new profiles
using these compounds are developed, and mechanism assignments for these
compounds will need to be made.
-
Poorly defined or complex mixture categories will need to
be removed as use of these categories are discontinued as part of ongoing
projects to improve existing emissions profiles. Ultimately, the only mixture
categories that should be retained are those necessitated by analytical
uncertainties.
-
The current database needs to be made available on the web
in such a manner that emissions profile developers can readily utilize
it to determine how to categorize the species in their profiles without
adding duplicate or incompatible entries.
The ultimate success of this project requires the widespread
adoption and use of this database (or one developed from it) when emissions
profiles are developed and updated in the future. This will require development
of standard procedures to update and add to the database as needed, and
central maintenance of the database by an appropriate and recognized authority.
Otherwise, the database will either not be used or will soon become outdated
and something resembling the current disorganized and inconsistent system
will evolve again.
References
CARB (2003): Organic Gas Speciation Profiles
provided by the California Air Resources Board at a web site that has been
moved. Downloaded file named ORGPROF_03_19_03.xls and dated 3/19/2003.
Carter, W. P. L. (2000a): "Documentation
of the SAPRC-99 Chemical Mechanism for VOC Reactivity Assessment," Report
to the California Air Resources Board, Contracts 92-329 and 95-308, May
8. Available at ../reactdat.htm.
Carter, W. P. L. (2000b): "Implementation
of the SAPRC-99 Chemical Mechanism into the Models-3 Framework," Report
to the United States Environmental Protection Agency, January 29. Available
at ../absts.htm#s99mod3.
Carter, W. P. L. (2007): "Development of the SAPRC-07 Chemical Mechanism
and Updated Ozone Reactivity Scales," Final report to the California Air
Resources Board Contract No. 03-318. August. Available at ../SAPRC.
Gipson (2001). PROFILE.VOC.DAT
file, used for processing VOC emissions data for Models-3. Received from
Gerald L. Gipson, EPA, Research Triangle Park, NC,. November, 2001
MCNC (2000): "Sparse Matrix Operator
Kernel Emissions (SMOKE) Modeling System,"
SPECIATE (2001): Speciate 2.1
database was available at http://www.epa.gov/ttn/chief/software/speciate/
as of late 2001. Data in spreadsheet format provided by Ronald Ryan, EPA,
Research Triangle Park, NC, November 2001. It has now been superseded by
the Speciate 3.2 database, which is now what
is available at that website.
Speciate (2002). Speciate 3.2 database available
at http://www.epa.gov/ttn/chief/software/speciate/
as of early 2004. Dated November 3, 2002.
Yarwood (2002). Texas Natural
Resource Conservation Commission emissions species data file "compound.database.eps2x.17May02",
provided by Greg Yarwood, Environ corporation, Novato, CA, Latest version
provided June 18, 2002
Allen (2001). California Air Resources
Board chemical species data file provided by Paul Allen, CARB, Sacramento,
CA, November 26, 2001. Superseded by the CARB (2003)
profiles.
Update History
| 2/10/14 | |
| 4/1/13 | - Assignments for MCM added to emitdb.xls.
- Errors in RACM2 assignments for nonvolatile mixtures were corrected.
|
| 3/26/13 | - The assigments for RACM2 have been updated to reflect the published
version of the mechanism. The assignments were reviewed and approved by
Wendy Goliff and Bill Stockwell, the developers of RACM2.
- Several minor corrections were made and a few minor new categories added to represent current SAPRC detailed model specie
|
12/18/11
|
- The assigments for RACM2 have been updated to reflect the current
version of the mechanism. The assignments were reviewed and approved by Bill Stockwell, one of the RACM2 developers..
- Revisions were made to Carbon Bond 05 assignments for several alkenes as recommended by the CB05 developers
|
| 4/28/11 | - The "Emit Worksheet" sheet of emitdb.xls
was modified to allow processing of additional mechanisms. No
assignments were changed and the normal processing procedures are not
affected
|
| 7/23/02 |
-
Some corrections made to SAROAD assignments and a few mixture assignments
-
"Denaturant" returned to a category by itself, and is again unassigned.
-
CO is removed from the list of SAPRC-99 lumped VOC categories, based on
the assumption that CO emissions are generally processed separately from
VOCs.
-
Option implemented to lump non-volatile compounds with unassigned compounds,
as requested by Greg Yarwood
- Option implemented to output names of database categories assigned
to SAROAD classes in output file giving lumped model species assignments
to SAROAD categories
|
| 7/2/02 |
-
Some SAPRC-99 lumped model species names changed to conform to requirements
for some models that they be no more than 4 characters
-
"Denaturant" assigned to methanol
- TAME given Texas SAROAD number of 99997 and CB4 splits for it changed
as Greg Yarwood recommended.
|
| 6/28/02 |
-
Output format of the lumped mechanism assignments for SAROAD categories
was modified. Now a molecular weight is assigned for each category where
that information is available, and the assignments are given in terms of
moles of model species per mole of category. (The molecular weight of mixtures
is defined as 1 gram / total number of moles of compounds assigned to the
mixture per gram, and the number of moles of the category is defined as
the number of moles of assigned compounds.) The ASCII file that can be
produced to give the lumped species assignments also now includes the molecular
weights assigned to the lumped classes.
-
Some minor corrections were made for some categories, and unused mixture
categories were removed.
-
A brief discussion of the formats of the mechanism assignment ASCII files
output by the macros was added to the "Documentation" sheet.
|
| 1/8/04 |
-
Many changes made to the speciation assignments as part of work carried
out during the previous 1 1/2 years, including work for the ongoing ACC
project. A number of errors corrected and other improvements made to the
assignments.
-
Macros were incorporated to output the assignment data into ASCII files
that can be read by the Fortran speciation processing programs being prepared
for the ACC project.
The profile database file was added to the distribution.
|
| 4/7/04 |
-
Changes made to the speciation assignments as part of the ongoing projects.
Several errors corrected and some additions made to the assignments.
-
Significant progress was made in preparing the emissions processing programs
for the ACC project. The preliminary documentation
for these programs was updated accordingly.
|
| 4/23/04 |
-
Assignments for RADM-2 mechanism added to the database file
-
Changes made to the SAPRC-99 emissions groups to accomodate future RACM
assignments
-
Corrections made to assignments for a few compounds.
|
| 5/17/04 |
-
Minor corrections made to "unknown" and "unassigned" profiles in profdb.xls
|
| 6/1/04 |
-
Corrected profile documentation for "base ROG" and "total emissions" profiles
in profdb.xls
|
| 6/7/04 |
-
Corrected bug in emitdb.xls macro ProcessSarEmit.
|
| 7/21/04 |
-
Profiles added to represent mixtures used in SAPRC-99 evaluation experiments
and for SAPRC-99 reactivity scales. Speciation and mixture categories used
in these profiles were added.
-
Errors were found in the SAPRC-99 mechanism files
in the preliminary program files (dated June 1, 2004) that affected reactions
generated by MechPro in the CMAQ (.MEC) format. (The names ISOPROD, METHACRO,
and PROD2 was used for IPRD, MACR, and PRD2 in some places in the generated
mechanism files.) These have been corrected. Mechanisms generated
using files downloaded previously should not be used.
|
| 12/22/04 |
-
Additional profiles added. These include petroleum distillate compositions
from the study of Cencullo et al (2002) and versions of EPA profiles currently
used by the University of Houston in Texas modeling studies.
-
Assignments were made to Texas "contaminant" code categories used in Texas
point source profiles.
-
Updates, corrections, and additions made to speciation processing programs.
-
Programs and examples prepared for processing EPS profiles in the Texas
database (preliminary)
|
| 1/17/05 |
-
The MechPro program updated to output a species table .CSV file for compiling
mechanisms for CMAQ. The program documentation was updated accordingly.
|
| 2/21/05 |
-
EMITROG1 profile added to serve as a base ROG mixture based on EPA emissions
data for relative reactivity calculations.
|
| 8/24/05 |
-
Speciation assignments were added to support profiles and new categories
in a preliminary version
of the Speciate 4.0 database.
-
Carbon Bond 4 assignments were modified for some compounds to improve self-consistency
among assignments.
-
Assignments were added for the new Carbon Bond '05 mechanism.
-
Model species assignments were made to a number of previously unassigned
compounds. Compounds are now either assigned for all supported mechanisms
or assigned for none.
-
A few additional profiles added to permit assignments of several complex
mixture categories.
-
Some modifications were made to the emitdb.xls spreadsheet.
-
The capability was added for the spreadsheet to assign model species to
unknown mixtures or unassigned compounds using profiles designed for this
purpose.
-
Some minor modifications were made to the programs. The major changes were
that the dimensions had to be increased to support the increased number
of speciation categories.
-
Molecular weights were modified slightly to be consistent with IUPAC atomic
weights. The largest atomic weight change is about 0.006%.
|
| 8/30/05 |
-
Minor corrections made to speciation assignments
|
| 7/26/06 |
-
Carbon Bond 4 and Carbon Bond '05 assignments were modified based on recommendations
by Uarporn Nopmongcol, Greg Yarwood and Gary Z. Whitten of Environ to Mark
Houyoux of the EPA dated May 9, 2006
-
A NVOL (nonvolatile) model species added for all lumped mechanisms and
used to represent non-volatile compounds. For CB4 and CB05 this is number
of non-volatile carbons. For Lumped Molecule mechanisms this kg non-volatile
mass (i.e., as if NVOL has a molecular weight of 1000). (Note that this
assignment is not made for uncharacterized non-volatile mixtures.)
-
The Carbon number and molecular weight for the SAPRC-99 INERT model species
were modified to reflect the mixture of inert compounds in the EMITBAS1
mixture (Profile representing total anthropogenic emissions obtained from
EPA Models-3 emissions databases (EPA, 1998), with methane, unknowns, nonvolatiles
and negligible contribution compounds removed.)
-
Went back to using Access internal ID numbers for Speciate 4 categories
-
References to Emitdb macros whose accuracy could not be assured, and cells
that they update, have been removed. Model species assignments are no longer
given on the "Master List" and "Saroad Assignments" spreadsheet. Model
species assignments for compounds are given on the "compounds" and "Master
Assignments" sheets. Model species assignments for other categories are
given only on the "Master Assignments" list. (Most of these were replaced
in the 8/4/06 version.)
-
This version of EmitDB.xls was found to have errors in the "Master Assignments"
sheet and should be replaced by the 8/4 or a later version.
|
| 8/4/06 |
-
Errors in macros used to produce assignments the "Master Assignments" sheet
have been corrected.
-
Macros used to produce model species assignments in the "Master List" and
"Saroad Assignments" sheets have been restored because the errors have
been fixed. No change in the primary compound or mixture assignments.
|
| 8/5/06 |
-
Macros modified so that model species assignments in the "Saroad Assignments"
sheet are updated at the same time those in the "Master List" sheet are
calculated. No change in the assignment data.
|
| 8/7/06 |
-
Three new Speciate 4 categories added to the "Saroad Assignments" sheet.
|
| 8/10/06 |
-
Several previously unassigned Speciate 4 categories given compound, molecular
weight, and mechanism assignments.
|
| 8/12/06 |
-
Category type codes now indicate whether complex mixtures have been assigned
to known compounds or not.
-
Some new compounds added to the database on 8/10 to define simple mixtures
were not on the "master list" sheet in the previous database. This has
been corrected. Some other minor corrections made.
|
| 8/31/07 |
-
Assignments for the SAPRC-07 mechanism added to emitdb.xls. However, the
speciation programs have not been updated, nor have all the files needed
by MechPro been created for this mechanism. See ../SAPRC
for information about SAPRC-07.
-
Some new compounds added to compounds list. These are primarily compounds
added as part of the SAPRC-07 mechanism update.
-
Several new unspeciated mixture profiles, which are included in the reactivity
tabulation for SAPRC-07, have been added to profdb.
|
| 12/31/07 |
-
Emissions assignments and model species revised for modified SAPRC-07 amine
mechanism.
|
| 1/3/08 |
-
Error in SAPRC-99 assignment files caused with the SAPRC-07 (August 31,
2007) update corrected. (This affects only the the macros in emitdb.xls
and the ASCII files they create, not the assignments in emitdb.xls).
|
| 2/4/08 |
-
Error in assignment for propene in the RADM-2 mechanism has been corrected.
|
| 5/27/08 |
-
Some minor corrections made to some assignments and a few new compounds
added
-
Hydrocarbon volatility cutoff for hydrocarbons now strictly set at C20
for consistency.
-
Added assignments for a preliminary version of a "Toxics" SAPRC07 mechanism,
SAPRC07T, that is still under development
-
Added assignments for a preliminary version of a condensed SAPRC07 mechanism,
CS07, that is still under review
|
| 8/25/08 |
-
Preliminary assignments added for the RACM2 mechanism of Goliff and Stockwell.
This required adding new sheets for assignment data. These need to be checked
by the RACM2 mechanism developers and are therefore subject to change.
|
|
| 10/6/08 |
-
Emissions categories added to cover all current SAPRC-07 detailed model
species
-
Mixture used to represent aloft VOCs in reactivity scale calculations added
to profdb.xls
-
Emitdb.xls now outputs condensed mechanism lumping (LCC) files used by
SAPRC modeling software.
|
10/15/08,
10/29/08 |
-
Added several chemical categories new in Speciate 4.2
|
| 4/30/09 | - Added
MechPro input files for SAPRC-07 mechanism and updated SAPRC-07 to the
3/09 version. Corrected an error in MechPro in formating RXN output
files.
|
Database files
|
|
-
Profdb.xls (dated October 8, 2008) (Link deleted: out of date. Does not reflect updates for Speciate 4).
|
- SpTool.zip Speciation
Tool assignment files (CSV format). Link deleted; out of date. See home page for current assignment files.
|
Program
files, Documentation, and Reports
-
Final report to the ACC project
titled "Integration of the SAPRC Chemical Mechanism in the SMOKE
Emissions Processor for the CMAQ/Models-3 Airshed Model", dated mid-2005.
|
|
|
|
|
-
specpgms.zip Version of programs and input and output files as of updated April 30, 2008). See the
README.TXT file for more information. Superceded by SpecDB.zip.
- This also includes files for processing SAPRC-99 with benzene and 1,3-butadiene
explicit.
|
Related links
[top of page]

